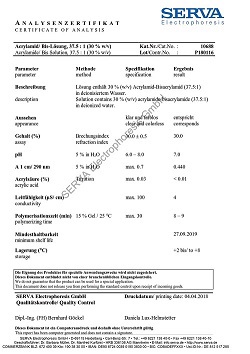
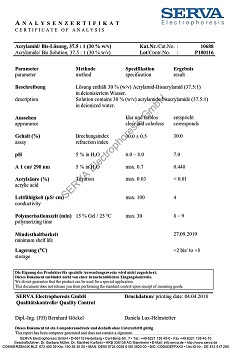
N-Lauroylsarcosine, Na-salt (30 % solution)
Sarcosyl surfactants can be considered as modified fatty acids, where the hydrocarbon chain is interrupted by an amidomethyl (-CONCH3-) group. The structure is similar to soaps, but due to the sarcosyl group, they show a better solubility in water and no sensitivity to calcium ions. The lauric acid in our product is of plant origin.
In biotechnology, N-Lauroylsarcosinate is mainly used for the isolation and characterization of DNA, proteins and enzymes, and for tissue homogenization. Diluted solutions of N-lauroylsarcosinate show at acid pH bacteriostatic properties.
Synonyms: N-Methyl-N-(oxododecyl)-glycine, sodium salt; Sarkosyl NL-30, Oramix L30
CAS registry number: [137-16-6]
Molecular Formula: C15H28NO3Na
Relative molecular mass: 293.4
Classification: Anionic surfactant
CMC: 13.7 mM
Bibliography
Extraction and Characterization of DNA and DNA Fragments
Hicks, K.E. et al. (1995) A simple and sensitive DNA hybridization assay used for the routine diagnosis of human Parvovirus B19 infection. J. Clin. Microbiol. 33, 2473-5.
Weiler, J. et al. (1997) Hybridisation based DNA screening on peptide nucleic acid (PNA) oligomer arrays. Nucl. Acids Res. 25, 2792-9.
Nakamura, Y. et al. (1999) Expression of soybean b-1,3-endoglucanase cDNA and effect on disease tolerance in kiwifruit plants. Plant Cell Rep. 18, 527-32.
Li, H. et al. (2001) A rapid and high yielding DNA miniprep for cotton (Gossypium spp.). Plant Mol. Biol. Rep. 19, 1-5.
Sun, L.V. et al. (2001) Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J. Bacteriol. 183, 2219-25.
Cosa, G. et al. (2002) DNA damage detection technique applying time-resolved fluorescence measurements. Anal. Chem. 74, 6163-9.
Isolation and Investigation of RNA
MacDonald, R.J. et al. (1987) Isolation of RNA using guanidinium salts. Meth. Enzymol. 152, 219-227.
Kimura, H. et al. (1999) Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 19, 5383-92.
Suzuki, F. et al. (2002) Distribution of alpha-1 adrenoceptor subtypes in RNA and protein in rabbit eyes. Brit. J. Pharmacol. 135, 600-8.
Pant, B.D. et al. (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150, 1541-55.
Isolation and Characterization of Enzymes and Proteins
Magnuson, T.S. et al. (2000) Characterization of a membrane-bound NADH-dependent Fe3+ reductase from the dissimilatory Fe3+-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol. Lett. 185, 205-11.
Bader, J.A. et al. (2004) Immune response induced by N-lauroylsarcosine extracted outer- membrane proteins of an isolate of Edwardsiella ictaluri in channel catfish. Fish Shellfish Immunol. 16, 415-28.
Kahlau, S. a. Bock, R. (2008) Plastid transcriptomics and translatomics of tomatoe fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20, 856-74.
Park, D.-W. et al. (2011) Improved recovery of active GST-fusion proteins from insoluble aggregates: solubilization and purification conditions using PKM2 and HtrA2 as model proteins. BMBRep.2011.44.4.279
Further Applications
Verbavatz, J.M. et al. (1993) Tetrameric assembly of CHIP28 water channels in liposomes and cell membranes: a freeze-fracture study. J. Cell Biol. 123, 605-18.
Rund, S. et al. (1999) Structural analysis of the lipopolysaccharide from Chlamydia trachomatis serotype L2. J. Biol. Chem. 274, 16819-24.
Piret, J. et al. (2002) Comparative study of mechanisms of Herpes simplex virus inactivation by sodium lauryl sulfate and N-lauroylsarcosine.
Langeveld, J.P.M. et al. (2003) Enzymatic degradation of prion protein in brain stem from infected cattle and sheep. J. Infect. Dis. 188, 1782-9.
Tamgüney, G. et al. (2009) Measuring prions by bioluminescence imaging. PNAS 106, 15002-6.